
Functional genomics of the Mycobacterium tuberculosis complex
We are studying the genetic and molecular basis of virulence in members of the Mycobacterium tuberculosis complex.
Challenge
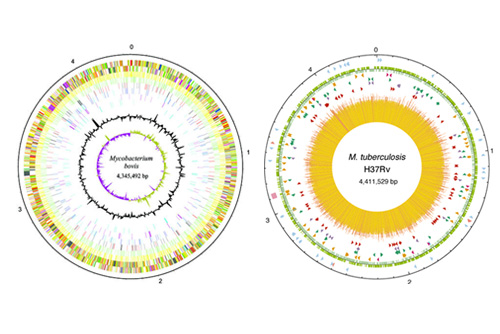
The Mycobacterium tuberculosis complex is comprised of a group of closely related bacteria with distinct host preferences. Species such as Mycobacterium bovis causes TB in animals, but has a wide host range. Species such as Mycobacterium tuberculosis and Mycobacterium africanum primarily cause TB in humans. Human TB is one of the top 10 causes of death from a single infectious agent and causes approximately 1.5 million deaths annually. The costs of control programmes of M. bovis infection in domestic livestock are prohibitively expensive and in LMIC M. bovis can represent a zoonotic risk. Unlike many other bacterial pathogens, members of the MTBC are highly genetically similar (>99% similar) and so we are trying to understand the genetic basis of host adaptation and virulence in both M. bovis and M. tuberculosis using a One Health approach. Despite the availability of many genome sequences we still do not understand the roles of many genes in virulence (e.g. virulence factors).
Solution
We are using whole genome approaches (Transposon insertion sequencing) to identify the gene required for survival in vivo. Additionally, we are using CRISPRi mediated gene silencing to investigate host pathogen interactions for selected gene targets. We are also utilising CRISPRi to determine target vulnerabilities for therapeutic agents for human tuberculosis. Through a better understanding of host:pathogen interactions in the two species we hope to generate novel therapeutics.
Partners
The work has been funded by several sources including; The Royal Society, The International Veterinary Vaccinology Network, the BBSRC and Defra
